Minus sediment concentration in river sediment case
Minus sediment concentration in river sediment case
Hello! I add two rivers in my model. When I check the sediment concentration, I found, near the river mouth, one river have normal sediment concentration and another always have minus sediment concentration. And both river have positive sediment mass. Is it a bug? The sediment concentration cannot be minus.
I can run sediment model without problem. Now I want to add river sediment source, so I add follow CPP options to add river sediment import.
#define RIVER_SEDIMENT
#define TS_PSOURCE
#define UV_PSOURCE
I set the sand concentration, sand fraction, sand mass equal zero in initial file, just see how river sediment is transported.
Thanks!
I can run sediment model without problem. Now I want to add river sediment source, so I add follow CPP options to add river sediment import.
#define RIVER_SEDIMENT
#define TS_PSOURCE
#define UV_PSOURCE
I set the sand concentration, sand fraction, sand mass equal zero in initial file, just see how river sediment is transported.
Thanks!
-
linzhenhua
- Posts: 64
- Joined: Mon Oct 17, 2005 2:02 am
- Location: Institute of Oceanology,Chinese Academy of Sciences
Re: Minus sediment concentration in river sediment case
Try to search "MPDATA" in the forum and check if the related discussions help.
Re: Minus sediment concentration in river sediment case
Dear linzhenhua,
I always used TS_MPDATA in my river model. But the sediment concentration is always minus near one river mouth. Thank you for your suggestion.
I always used TS_MPDATA in my river model. But the sediment concentration is always minus near one river mouth. Thank you for your suggestion.
Re: Minus sediment concentration in river sediment case
can u check if the flows are correctly coming out of the river? so look at the history file and compute the flow out of the river. Is the flow direction tidal or does it always flow into the grid?
Re: Minus sediment concentration in river sediment case
I check the flows at river point, it is tidal, sometimes flow into the grid, sometimes flow out. At first 2 hours the flow direction is flow into the grid, the concentration is 0. When flow direction change to flowing out, the concentration become minus and always be minus since then, no matter the flow direction flowing in or out.jcwarner wrote:can u check if the flows are correctly coming out of the river? so look at the history file and compute the flow out of the river. Is the flow direction tidal or does it always flow into the grid?
Re: Minus sediment concentration in river sediment case
this could be the problem: the way these point sources are coded in, i can not guarantee positivity of the sediment. Here is an example: IF the flow is out of the grid, and the flow is going to take more sediment out of a cell than actually exists in the cell, then the concentrations will go negative. The MPDATA scheme itself will prevent this from happening, since it is basically upwind with a higher order correction. Upwind will prevent negative values. BUT ---> the river sources(/sinks) are not time stepped inside the MPDATA loops. they are added as additional terms. So i have no control over them. This may be what is happening, i am not sure. One way to test this is to not take any sed when the flow is out of the grid. So a test would be "if Qriver is out of hte grid, then set sed_river=0" This is only a test - to identify the problem. it is not the solution.
Re: Minus sediment concentration in river sediment case
Dear Warner, thanks for your advice. I am confused about this. I have 3 kind of sediment in my model(sand_01,sand_02,sand_03). I set sand_01 sediment mass equal zero in all cells in initial file. So there doesn't exist sand_01 sediment at any cell. All the sand_01 sediment come from the source point(river point). I thought the river point should have infinite sediment source. If not, I need set a very very huge sediment mass at river point cell to make sure there is enough sediment to be flowed out? This also make no sense, because another river in my model have a positive concentration. Is there something I misunderstanding?jcwarner wrote: Here is an example: IF the flow is out of the grid, and the flow is going to take more sediment out of a cell than actually exists in the cell, then the concentrations will go negative.
-
coelho
- Posts: 7
- Joined: Fri Feb 13, 2009 7:39 pm
- Location: Oceanographic Institute of Sao Paulo University
Re: Minus sediment concentration in river sediment case
Did you check if your river is located beside a land point? Maybe the negative sediment values are because of that.
Re: Minus sediment concentration in river sediment case
The river is located beside two land points. Did you mean the river should not near to land point? But another river is located beside three land points, its concentration is positive.coelho wrote:Did you check if your river is located beside a land point? Maybe the negative sediment values are because of that.
-
coelho
- Posts: 7
- Joined: Fri Feb 13, 2009 7:39 pm
- Location: Oceanographic Institute of Sao Paulo University
Re: Minus sediment concentration in river sediment case
I mean that the river should be near to land point. I had some problems with rivers located not exactly beside a land point. Try to move the location of the river that you are getting negative values and see what happens.CBian wrote: The river is located beside two land points. Did you mean the river should not near to land point? But another river is located beside three land points, its concentration is positive.
Re: Minus sediment concentration in river sediment case
so which of the 3 sediments go negative?
please explain your initial setup for all 3 seds, and how many come in through the river, and what the time series of sed for the river input is.
please explain your initial setup for all 3 seds, and how many come in through the river, and what the time series of sed for the river input is.
Re: Minus sediment concentration in river sediment case
Thanks, coelho. Yes, I always make my river point next to land point. I tried to move the river locations and river directions, the river always have negative values no matter whatever I did.coelho wrote:
I mean that the river should be near to land point. I had some problems with rivers located not exactly beside a land point. Try to move the location of the river that you are getting negative values and see what happens.
Re: Minus sediment concentration in river sediment case
I expended my model domain to include more area. However, in my new model case (still include above two rivers), the river have no influence on the model result. It seems no river added to the model. The salt, temperature, sediment concentration have no difference with the model results without river input.
This is very strange. I check the output log file.
T LtracerSrc(01) Processing point sources/Sink on tracer 01: temp
T LtracerSrc(02) Processing point sources/Sink on tracer 02: salt
T LtracerSrc(03) Processing point sources/Sink on tracer 03: sand_01
T LtracerSrc(04) Processing point sources/Sink on tracer 04: sand_02
T LtracerSrc(05) Processing point sources/Sink on tracer 05: sand_03
GET_NGFLD - river runoff XI-positions at RHO-points
(Min = 3.40000000E+01 Max = 3.40000000E+01)
GET_NGFLD - river runoff ETA-positions at RHO-points
(Min = 7.20000000E+01 Max = 7.20000000E+01)
GET_NGFLD - river runoff direction
(Min = 0.00000000E+00 Max = 0.00000000E+00)
GET_NGFLD - river runoff mass transport vertical profile
(Min = 0.00000000E+00 Max = 2.00000000E-01)
GET_NGFLD - river runoff mass transport, t = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 1.04000000E+04 Max = 1.04000000E+04)
GET_NGFLD - river runoff potential temperature, t = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 4.00000000E+00 Max = 4.00000000E+00)
GET_NGFLD - river runoff salinity, t = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 4.00000000E+00 Max = 4.00000000E+00)
GET_NGFLD - river runoff suspended sediment concentratit = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 2.23308394E-01 Max = 2.23308394E-01)
GET_NGFLD - river runoff suspended sediment concentratit = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 1.33985037E-01 Max = 1.33985037E-01)
GET_NGFLD - river runoff suspended sediment concentratit = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 0.00000000E+00 Max = 0.00000000E+00)
It seems the river data were used by model. But why the result have no different at all? I don't know where I changed which induced this problem. Can anyone give me some advice?
This is very strange. I check the output log file.
T LtracerSrc(01) Processing point sources/Sink on tracer 01: temp
T LtracerSrc(02) Processing point sources/Sink on tracer 02: salt
T LtracerSrc(03) Processing point sources/Sink on tracer 03: sand_01
T LtracerSrc(04) Processing point sources/Sink on tracer 04: sand_02
T LtracerSrc(05) Processing point sources/Sink on tracer 05: sand_03
GET_NGFLD - river runoff XI-positions at RHO-points
(Min = 3.40000000E+01 Max = 3.40000000E+01)
GET_NGFLD - river runoff ETA-positions at RHO-points
(Min = 7.20000000E+01 Max = 7.20000000E+01)
GET_NGFLD - river runoff direction
(Min = 0.00000000E+00 Max = 0.00000000E+00)
GET_NGFLD - river runoff mass transport vertical profile
(Min = 0.00000000E+00 Max = 2.00000000E-01)
GET_NGFLD - river runoff mass transport, t = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 1.04000000E+04 Max = 1.04000000E+04)
GET_NGFLD - river runoff potential temperature, t = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 4.00000000E+00 Max = 4.00000000E+00)
GET_NGFLD - river runoff salinity, t = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 4.00000000E+00 Max = 4.00000000E+00)
GET_NGFLD - river runoff suspended sediment concentratit = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 2.23308394E-01 Max = 2.23308394E-01)
GET_NGFLD - river runoff suspended sediment concentratit = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 1.33985037E-01 Max = 1.33985037E-01)
GET_NGFLD - river runoff suspended sediment concentratit = 345 00:00:00
(Rec=0012, Index=1, File: byecs-river.nc)
(Tmin= 15.0000 Tmax= 345.0000)
(Min = 0.00000000E+00 Max = 0.00000000E+00)
It seems the river data were used by model. But why the result have no different at all? I don't know where I changed which induced this problem. Can anyone give me some advice?
-
coelho
- Posts: 7
- Joined: Fri Feb 13, 2009 7:39 pm
- Location: Oceanographic Institute of Sao Paulo University
Re: Minus sediment concentration in river sediment case
The Min and Max values for XI-positions and ETA-positions are the same. Are you sure you are using two rivers?CBian wrote: GET_NGFLD - river runoff XI-positions at RHO-points
(Min = 3.40000000E+01 Max = 3.40000000E+01)
GET_NGFLD - river runoff ETA-positions at RHO-points
(Min = 7.20000000E+01 Max = 7.20000000E+01)
Can you send your byecs-river.nc file and your grid file?
Re: Minus sediment concentration in river sediment case
Yes, you are right, I made a mistake when I created the new river forcing file. Now I changed it and using two rivers. However, the rivers still have no influence.coelho wrote:The Min and Max values for XI-positions and ETA-positions are the same. Are you sure you are using two rivers?CBian wrote: GET_NGFLD - river runoff XI-positions at RHO-points
(Min = 3.40000000E+01 Max = 3.40000000E+01)
GET_NGFLD - river runoff ETA-positions at RHO-points
(Min = 7.20000000E+01 Max = 7.20000000E+01)
Can you send your byecs-river.nc file and your grid file?
I attached the grid and river file. Thanks!
- Attachments
-
- byecs-river.nc
- (22.36 KiB) Downloaded 2208 times
-
- byecs-grid.nc
- (3.42 MiB) Downloaded 1825 times
-
coelho
- Posts: 7
- Joined: Fri Feb 13, 2009 7:39 pm
- Location: Oceanographic Institute of Sao Paulo University
Re: Minus sediment concentration in river sediment case
Seems that everything is fine. Try to put all the 20 levels of river_Vshape = 0.05(attached) and see if you find any sand coming from the rivers.
Your river_sand_03 is always zero. Is that correct?
Your river_sand_03 is always zero. Is that correct?
- Attachments
-
- byecs-river_new.nc
- (22.36 KiB) Downloaded 1967 times
Re: Minus sediment concentration in river sediment case
There are 3 kind of sand in my model. I just 2 kind of sand from rivers, the third kind of sand doesn't come from rivers. So I set river_sand_03 zero.coelho wrote:Seems that everything is fine. Try to put all the 20 levels of river_Vshape = 0.05(attached) and see if you find any sand coming from the rivers.
Your river_sand_03 is always zero. Is that correct?
I tried your new river forcing file and found that there is sand coming from the river. I tried my old river forcing again, and found there is also sand from the river. But I paid too much attention to the Salt and Temp so that I didn't notice there is sand coming from river.
However, there still have problems.
First, one river output sand, but another river didn't.
Second, both rivers seem no influence on TS.
To sum up, it looks like just one river exist, another disappeared.
Third, the river which output sand is the river with negative sediment concentration river, it still make negative sediment concentration.
-
coelho
- Posts: 7
- Joined: Fri Feb 13, 2009 7:39 pm
- Location: Oceanographic Institute of Sao Paulo University
Re: Minus sediment concentration in river sediment case
Very odd.CBian wrote: There are 3 kind of sand in my model. I just 2 kind of sand from rivers, the third kind of sand doesn't come from rivers. So I set river_sand_03 zero.
I tried your new river forcing file and found that there is sand coming from the river. I tried my old river forcing again, and found there is also sand from the river. But I paid too much attention to the Salt and Temp so that I didn't notice there is sand coming from river.
However, there still have problems.
First, one river output sand, but another river didn't.
Second, both rivers seem no influence on TS.
To sum up, it looks like just one river exist, another disappeared.
Third, the river which output sand is the river with negative sediment concentration river, it still make negative sediment concentration.
I will run your domain just with river forcing and see if I find the same results.
-
coelho
- Posts: 7
- Joined: Fri Feb 13, 2009 7:39 pm
- Location: Oceanographic Institute of Sao Paulo University
Re: Minus sediment concentration in river sediment case
Hey,
I had run your model and I have got the same problem of negative sediment values but I got TS influence from one river.

The thing is that your model domain is very huge. I don't know if this is really the problem but your Dx and Dy is 18 km. My suggestion is to refine your grid at least in the rivers area.
Good luck.
I had run your model and I have got the same problem of negative sediment values but I got TS influence from one river.

The thing is that your model domain is very huge. I don't know if this is really the problem but your Dx and Dy is 18 km. My suggestion is to refine your grid at least in the rivers area.
Good luck.
Re: Minus sediment concentration in river sediment case
The scale you are working at seems to be very large. so you are not really resolving the dynamics at the river mouths. One way to get around this is to unmask one or 2 cells so that you can move the river input slightly landward to a location where the flow is always into the grid. This will help to remove the possibility for negative sediment, as the flow will always be into the grid.
Re: Minus sediment concentration in river sediment case
Hi, I refined my domain, and the Dx, Dy are changed from 18km to 5km. I didn't add sediment to the model this time, just want to see the salt result near river mouth.
Both of two rivers can be found this time. However, one water cell next to the river cell always has a largest salt value(salt >100). I have changed the river position several times, but the abnormal salt value always appeared next to river point. I use TS_MPDATA in my model. Why this always happened?
Both of two rivers can be found this time. However, one water cell next to the river cell always has a largest salt value(salt >100). I have changed the river position several times, but the abnormal salt value always appeared next to river point. I use TS_MPDATA in my model. Why this always happened?
Re: Minus sediment concentration in river sediment case
is the river flow coming into the model or out of the model?
Re: Minus sediment concentration in river sediment case
my river_direction is 0. I add tide in the model.jcwarner wrote:is the river flow coming into the model or out of the model?
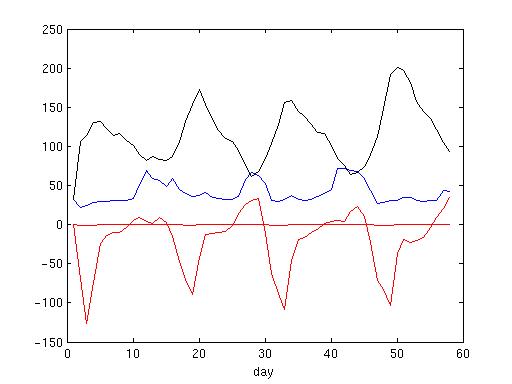
I output history result 24 hours a time.
The red line is the u*100 m/s. In my model, the left side of the river point is land, so there are strong flow coming into river, weak flow coming out of the river.
The blue line is the salt value at river cell. When river flow coming out, the salt could be more than 50.
The black line is the salt value next to river cell.
Re: Minus sediment concentration in river sediment case
is this flow correct? it seems that the net flow would be out of the domain, into the river. A
Re: Minus sediment concentration in river sediment case
jcwarner wrote:is this flow correct? it seems that the net flow would be out of the domain, into the river. A
In my opinion, the flow is correct. Because I check the current and tide harmonic constant results in this domain, the result looks well.
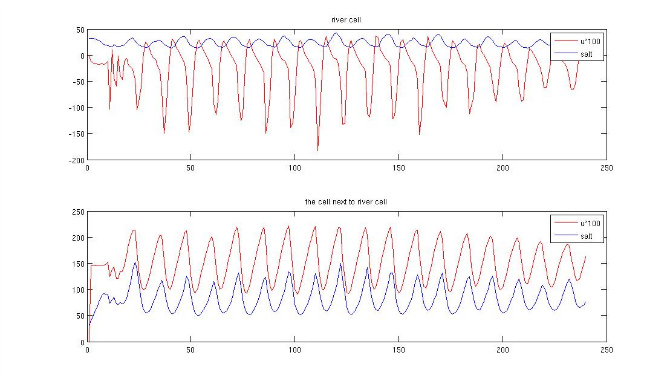
I tried to delete SCORRECTION cpp opinion in the model. I found the result look a little better. The salt in river cell looks normal, around 34. However, the cell next to river cell still have more than 100 value. And I check the u speed at this cell, the flow always flow into domain.
Re: Minus sediment concentration in river sediment case
I change the river position to the cell which has abnormal large salt. The result shows that this cell(now the river cell) have normal salt value, however, the cell next to this cell have more than 100 salt value. So abnormal salt always appeared next to the river cell. 
Re: Minus sediment concentration in river sediment case
i am still not clear about the direction of the river flow. If you set direction = 0, this just means it is in the XI ('x') direction. The magnitude of the flow can be positive or negative, which makes the transport be to the left or to the right. If you are specifying flow at a u point, and the flow value is negative, then the transport is to the left. You said that to the left of this point is land. So is the river flowing out of the domain?
maybe you could zoom in on a pcolor of salt with velocity arrows.
maybe you could zoom in on a pcolor of salt with velocity arrows.
Re: Minus sediment concentration in river sediment case
Hi Bian,
From my point of view, the abnormal "salinity" problem is also largely related to the grid or the land-sea mask at the river points. Plz also try to modify your "mask" and make the river more "smooth" or you may want to further dig the river land-ward straightly, and artificially give more grids to this "fake river" as well as larger water depth.
Although dynamically un-realistic, this may help you, if you are not concentrating on the estuarine circulations.
By the way, is "coelho" showing your model domain? if it is, it looks not a good model domain for ECSs studies, especially when you further invite the Kuroshio into consideration.
Just for your reference.
regards,
ZQ
From my point of view, the abnormal "salinity" problem is also largely related to the grid or the land-sea mask at the river points. Plz also try to modify your "mask" and make the river more "smooth" or you may want to further dig the river land-ward straightly, and artificially give more grids to this "fake river" as well as larger water depth.
Although dynamically un-realistic, this may help you, if you are not concentrating on the estuarine circulations.
By the way, is "coelho" showing your model domain? if it is, it looks not a good model domain for ECSs studies, especially when you further invite the Kuroshio into consideration.
Just for your reference.
regards,
ZQ
Re: Minus sediment concentration in river sediment case
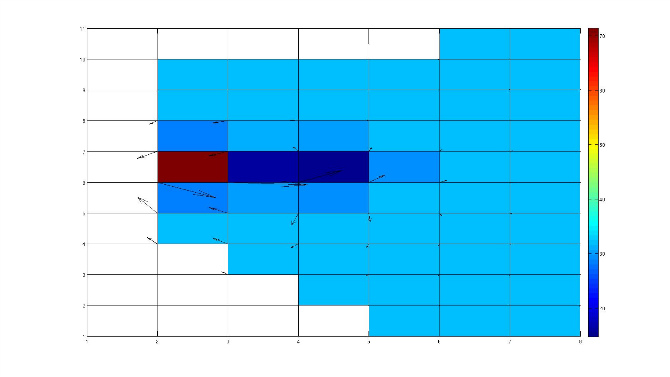
I plot the salt and velocity near river mouth. Yes, the river flows out of the domain. At the below figure, (5,2)is the river cell, (6,2) is the cell with abnormal salt (>70)jcwarner wrote:i am still not clear about the direction of the river flow. If you set direction = 0, this just means it is in the XI ('x') direction. The magnitude of the flow can be positive or negative, which makes the transport be to the left or to the right. If you are specifying flow at a u point, and the flow value is negative, then the transport is to the left. You said that to the left of this point is land. So is the river flowing out of the domain?
maybe you could zoom in on a pcolor of salt with velocity arrows.
Re: Minus sediment concentration in river sediment case
Very strange!! ZQ told me to move river point seaward to see what will happen. I tried it, same abnormal salt result. However, I tried to move river point landward. Now the model runs nearly one month, the salt is good and the river flow into sea too. But this means I put the river cell in land. I recheck the MASK file, mask_rho,mask_u are all land point in this cell.
-
coelho
- Posts: 7
- Joined: Fri Feb 13, 2009 7:39 pm
- Location: Oceanographic Institute of Sao Paulo University
Re: Minus sediment concentration in river sediment case
Hi CBian,CBian wrote:Very strange!! ZQ told me to move river point seaward to see what will happen. I tried it, same abnormal salt result. However, I tried to move river point landward. Now the model runs nearly one month, the salt is good and the river flow into sea too. But this means I put the river cell in land. I recheck the MASK file, mask_rho,mask_u are all land point in this cell.
Did you try to put any sediment with the new river location?
Re: Minus sediment concentration in river sediment case
the term 'river cell' is confusing. The rivers are a point source, so they are from a point in the model, not a cell. The point can (should) be at a location where the land mask (u_mask or v_mask) is 0. This will make the river come from the "land". I say u_mask or v_mask, because the river is specified at a u or v point, not a rho point.
Re: Minus sediment concentration in river sediment case
I still didn't put sediment into river, but I will do it as soon as possible and see if I still get negetive concentration near river mouth.coelho wrote:Hi CBian,
Did you try to put any sediment with the new river location?
I am very glad to know the river point can be located at "land".
Re: Minus sediment concentration in river sediment case
Hi Bian,
I want to remind you that the grid num. of ROMS should be started from 0, instead of 1 you read in the matlab maxtrix (Array Editor), which means you may already put the river correctly this time, even it looks like on the "land".
regards,
ZQ
I want to remind you that the grid num. of ROMS should be started from 0, instead of 1 you read in the matlab maxtrix (Array Editor), which means you may already put the river correctly this time, even it looks like on the "land".
regards,
ZQ
Re: Minus sediment concentration in river sediment case
I still cannot understand how the river location was set in the model. So I tried to put all the rivers in my domain to the model. There are 10 rivers in this area. After lots of tests, I added 7 of them to the model successfully. However, other 3 rivers cannot be accepted near their location. These 3 rivers always have abnormal salt value >100, or disappear.
I read a message from kate:
The results are very interesting, all the rivers can be successfully added in the model have follow characters:
mask_rho(lat,lon-1)==0 & mask_rho(lat,lon)==0 & mask_rho(lat,lon+1)==1
mask_u(lat,lon-1)==0 & mask_u(lat,lon)==0 & mask_u(lat,lon+1)==1
mask_v(lat,lon-1)==0 & mask_v(lat,lon)==0 & mask_v(lat,lon+1)==1
So I output all the points in my domain which have these characters and test some of them. All of them can be added as a river successfully. And if only two of mask_rho, mask_u, mask_v comply with up characters, the river also cannot be added.
But these case is applied for the river flow from west to east. If I have a river flow east to west, there will no point can have characters like:
mask_rho(lat,lon-1)==1 & mask_rho(lat,lon)==0 & mask_rho(lat,lon+1)==0
mask_u(lat,lon-1)==1 & mask_u(lat,lon)==0 & mask_u(lat,lon+1)==0
mask_v(lat,lon-1)==1 & mask_v(lat,lon)==0 & mask_v(lat,lon+1)==0
Because the mask_u are calculated like this:
[Mp,Lp]=size(mask_rho);
M=Mp-1;
L=Lp-1;
mask_u=mask_rho(:,1:L).*mask_rho(:,2:Lp);
so if mask_rho(lat,lon-1)==1 & mask_rho(lat,lon)==0, mask_u(lat,lon-1)must be 0 instead of 1.
I also tried lots of points, I cannot add river which flows from east to west.
And there are two rivers in model domain which flow from north to south, I also cannot add them in model.
So, is there anyone who can add a river flow from east to west and the river not disappear and the salt value also ok ?? Or the river can be added successfully without the characters mentioned above?
Any suggestion will be appreciated!
I read a message from kate:
So I test the mask_rho, mask_u, mask_v in the river point.kate wrote: If a source goes east to west, its indices will be at a place with zero mask_u, since that is the land-sea boundary. In fact, any source should be where the mask_u or mask_v is zero, with land on one side and water on the other.
The results are very interesting, all the rivers can be successfully added in the model have follow characters:
mask_rho(lat,lon-1)==0 & mask_rho(lat,lon)==0 & mask_rho(lat,lon+1)==1
mask_u(lat,lon-1)==0 & mask_u(lat,lon)==0 & mask_u(lat,lon+1)==1
mask_v(lat,lon-1)==0 & mask_v(lat,lon)==0 & mask_v(lat,lon+1)==1
So I output all the points in my domain which have these characters and test some of them. All of them can be added as a river successfully. And if only two of mask_rho, mask_u, mask_v comply with up characters, the river also cannot be added.
But these case is applied for the river flow from west to east. If I have a river flow east to west, there will no point can have characters like:
mask_rho(lat,lon-1)==1 & mask_rho(lat,lon)==0 & mask_rho(lat,lon+1)==0
mask_u(lat,lon-1)==1 & mask_u(lat,lon)==0 & mask_u(lat,lon+1)==0
mask_v(lat,lon-1)==1 & mask_v(lat,lon)==0 & mask_v(lat,lon+1)==0
Because the mask_u are calculated like this:
[Mp,Lp]=size(mask_rho);
M=Mp-1;
L=Lp-1;
mask_u=mask_rho(:,1:L).*mask_rho(:,2:Lp);
so if mask_rho(lat,lon-1)==1 & mask_rho(lat,lon)==0, mask_u(lat,lon-1)must be 0 instead of 1.
I also tried lots of points, I cannot add river which flows from east to west.
And there are two rivers in model domain which flow from north to south, I also cannot add them in model.
So, is there anyone who can add a river flow from east to west and the river not disappear and the salt value also ok ?? Or the river can be added successfully without the characters mentioned above?
Any suggestion will be appreciated!
Re: Minus sediment concentration in river sediment case
I can have rivers flowing in any direction. It's probably a question of your indices. In the ROMS manual, there are diagrams like:
with u-indices starting at 1 and h/rho indices starting at 0.
But in matlab/python/NCL, the indices will all start at 0 (or 1) and be more like:For this to be a west-flowing river, rmask(i+1)=0, rmask(i)=1, and umask(i)=0. Clear as mud?
Code: Select all
______|_______
|
h u h
|
______|_______
i-1 i iBut in matlab/python/NCL, the indices will all start at 0 (or 1) and be more like:
Code: Select all
______|_______
|
h u h
|
______|_______
i i i+1Re: Minus sediment concentration in river sediment case
Thanks, kate, you said very clear. But my result is different with what you said.
mathieu said the river_transport is negative when river flows westward or southward. After change the sign, I can add all river successfully.
However, one of my river is southward flow. I set the river point at river forcing file: Eposition=422, Xposition=118. jcwarner said before: the river is specified at a u or v point, not a rho point. So mask_v(422,118) should be 0, am I right? I use Matlab to check the result, so the v point 422 in Matlab also is 422. However, the mask_v(422,118)=1. This means the river is added in a wet point. But the model runs very well: the salt of the river mouth is decreasing. Is this means the river can also be added in some place with mask_v=1?
Even the model can run very well now, I still worry there will be some negative influence on the result.
mathieu said the river_transport is negative when river flows westward or southward. After change the sign, I can add all river successfully.
However, one of my river is southward flow. I set the river point at river forcing file: Eposition=422, Xposition=118. jcwarner said before: the river is specified at a u or v point, not a rho point. So mask_v(422,118) should be 0, am I right? I use Matlab to check the result, so the v point 422 in Matlab also is 422. However, the mask_v(422,118)=1. This means the river is added in a wet point. But the model runs very well: the salt of the river mouth is decreasing. Is this means the river can also be added in some place with mask_v=1?
Even the model can run very well now, I still worry there will be some negative influence on the result.
Re: Minus sediment concentration in river sediment case
Hi, coelho,coelho wrote:Hi CBian,CBian wrote:Very strange!! ZQ told me to move river point seaward to see what will happen. I tried it, same abnormal salt result. However, I tried to move river point landward. Now the model runs nearly one month, the salt is good and the river flow into sea too. But this means I put the river cell in land. I recheck the MASK file, mask_rho,mask_u are all land point in this cell.
Did you try to put any sediment with the new river location?
I put sediment with new river location, the sediment concentration seems normal, no minus concentration anymore, thanks!
-
ShinerM
Re: Minus sediment concentration in river sediment case
Hi,
I have met the same problems as yours when i added two rivers into my roms model.These two rivers flow from north to south. I set my river points next to the land point,however the salt value in the river point has the largest value(salt>128).And when I changed the river position to the land point which next to the water point,the salt value turned to be normal,but the velocity in the V direction in the river point(just the point i set my river entrance) are all zero.These river points are all in the land point.I wonder about all these,and now I don't know what is the problem and how to meet it.
Fortunately, I read your case when i am powerless recently.Would you tell me how did you solved your problems.Many thanks~
I am looking forward to your help.
I have met the same problems as yours when i added two rivers into my roms model.These two rivers flow from north to south. I set my river points next to the land point,however the salt value in the river point has the largest value(salt>128).And when I changed the river position to the land point which next to the water point,the salt value turned to be normal,but the velocity in the V direction in the river point(just the point i set my river entrance) are all zero.These river points are all in the land point.I wonder about all these,and now I don't know what is the problem and how to meet it.
Fortunately, I read your case when i am powerless recently.Would you tell me how did you solved your problems.Many thanks~
I am looking forward to your help.
Re: Minus sediment concentration in river sediment case
The river guidelines are posted here. Can you provide a drawing of the land mask around your river point and the i,j values?
-
ShinerM
Re: Minus sediment concentration in river sediment case
Thanks so much for your responses.kate wrote:The river guidelines are posted here. Can you provide a drawing of the land mask around your river point and the i,j values?
The files I attached are the river points in the mask_v from my grid file.The river points in the mask_v of the north to south river are (4,3)(5,3)(6,3),so the real river points I should get for the roms running are (3,3)(4,3)(5,3).
So as to the south to north river.The river points in the mask_v are (3,132)(4,132)(5,132),so the real river points I should get for the roms running are (2,132)(3,132)(4,132).Am I right?
When I set these river points in my make_forcing_river.m file,and the result of the roms runnning are strange.the salt value in the river points are all larger than 128.Both rivers have this case.
However,if i set the river points in (3,2)(4,2)(5,2),and (2,133)(3,133)(4,133)(these points are all land points),the salt value seems regular.but the velocity in the river points are all zero.
Waiting for your responses.Thanks.
Re: Minus sediment concentration in river sediment case
As I said back in March 2011, it depends on how you do your counting. Assuming you are counting in the Matlab/Python/ncview way, the indices of the v-points should match those of the rho-points for the north-to-south rivers and should be one less in j from the rho-points for the south-to-north rivers. In both cases, the i-indices should match those of the rho-points.ShinerM wrote:Thanks so much for your responses.kate wrote:The river guidelines are posted here. Can you provide a drawing of the land mask around your river point and the i,j values?
The files I attached are the river points in the mask_v from my grid file.The river points in the mask_v of the north to south river are (4,3)(5,3)(6,3),so the real river points I should get for the roms running are (3,3)(4,3)(5,3).
So as to the south to north river.The river points in the mask_v are (3,132)(4,132)(5,132),so the real river points I should get for the roms running are (2,132)(3,132)(4,132).Am I right?
When I set these river points in my make_forcing_river.m file,and the result of the roms runnning are strange.the salt value in the river points are all larger than 128.Both rivers have this case.
However,if i set the river points in (3,2)(4,2)(5,2),and (2,133)(3,133)(4,133)(these points are all land points),the salt value seems regular.but the velocity in the river points are all zero.
Waiting for your responses.Thanks.
Are you providing tracer values on inflow? If not, you are facing the dread down-stream advection scheme, known to be unstable.
-
machaielder
- Posts: 43
- Joined: Wed Mar 18, 2015 3:40 pm
- Location: Universidade Federal Fluminense
Re: Minus sediment concentration in river sediment case
Dear all
I am sorry for catting, I think this is an ideal discussion to clear out my doubts.
My aim with the model is simulate suspended sediments from River interring the shelf and I am still getting familiar with the model.
I firs run the upwelling example to test my roms and it runs ok.
Now I added the bathymetry (GRDNAME) and made some definitions;
When I define define SALINITY #define SOLVE3D #define SPLINES it gives me the error below (INPUT 1).
when I don’t define those parameters it runs but the value of outputs does not vary with time-steps.
I have download salinity, temperature and tide data from WOAD09 and TPX07.1 respectively to force the model but I don't know how to include set these forcings.
If You don’t mind I would like to know the settings I should make to configure the model for sediments.
Thank you in advance
Helder
INPUT 1
** Application flag: SOFALA
** Input script: sofala.in
*/
#define UV_ADV
#define UV_COR
#define UV_QDRAG
#define UV_VIS2
#define MIX_S_UV
#define UV_C4ADVECTION
#define SALINITY
#define SOLVE3D
#define SPLINES
#define MASKING
#define SPLINES
#define MY25_MIXING
#ifdef MY25_MIXING
# define N2S2_HORAVG
# define KANTHA_CLAYSON
#endif
#define DJ_GRADPS
#define ANA_SMFLUX
#define ANA_INITIAL
#define ANA_FSOBC
#define ANA_M2OBC
OUTPUT
Nonlinear model elapsed time profile:
Allocation and array initialization .............. 0.513 (61.3738 %)
2D/3D coupling, vertical metrics ................. 0.094 (11.2055 %)
Omega vertical velocity .......................... 0.035 ( 4.2247 %)
Equation of state for seawater ................... 0.095 (11.3984 %)
Total: 0.737 88.2024
All percentages are with respect to total time = 0.836
ROMS/TOMS - Output NetCDF summary for Grid 01:
Analytical header files used:
ROMS/Functionals/ana_initial.h
ROMS/TOMS - Output error ............ exit_flag: 3
ERROR: Abnormal termination: NetCDF OUTPUT.
REASON: NetCDF: Unknown file format
...........................................................................................
..............................................................................................
INPUT EXAMPLE 2
#define UV_ADV
#define UV_COR
#define UV_QDRAG
#define UV_VIS2
#define MIX_S_UV
#define UV_C4ADVECTION
#define MASKING
#define SPLINES
#define MY25_MIXING
#ifdef MY25_MIXING
# define N2S2_HORAVG
# define KANTHA_CLAYSON
#endif
#define DJ_GRADPS
#define ANA_SMFLUX
#define ANA_INITIAL
#define ANA_FSOBC
#define ANA_M2OBC
OUTPUT 2
STEP Day HH:MM:SS KINETIC_ENRG POTEN_ENRG TOTAL_ENRG NET_VOLUME
C => (i,j) Cu Cv C Max Max Speed
0 0 00:00:00 0.000000E+00 0.000000E+00 0.000000E+00 1.403707E+15
(000,000) 0.000000E+00 0.000000E+00 0.000000E+00 0.000000E+00
DEF_HIS - creating history file, Grid 01: sofala_his.nc
WRT_HIS - wrote history fields (Index=1) into time record = 0000001
1 0 00:05:00 0.000000E+00 0.000000E+00 0.000000E+00 1.403707E+15
(000,000) 0.000000E+00 0.000000E+00 0.000000E+00 0.000000E+00
2 0 00:10:00 0.000000E+00 0.000000E+00 0.000000E+00 1.403707E+15
(000,000) 0.000000E+00 0.000000E+00 0.000000E+00 0.000000E+00
I am sorry for catting, I think this is an ideal discussion to clear out my doubts.
My aim with the model is simulate suspended sediments from River interring the shelf and I am still getting familiar with the model.
I firs run the upwelling example to test my roms and it runs ok.
Now I added the bathymetry (GRDNAME) and made some definitions;
When I define define SALINITY #define SOLVE3D #define SPLINES it gives me the error below (INPUT 1).
when I don’t define those parameters it runs but the value of outputs does not vary with time-steps.
I have download salinity, temperature and tide data from WOAD09 and TPX07.1 respectively to force the model but I don't know how to include set these forcings.
If You don’t mind I would like to know the settings I should make to configure the model for sediments.
Thank you in advance
Helder
INPUT 1
** Application flag: SOFALA
** Input script: sofala.in
*/
#define UV_ADV
#define UV_COR
#define UV_QDRAG
#define UV_VIS2
#define MIX_S_UV
#define UV_C4ADVECTION
#define SALINITY
#define SOLVE3D
#define SPLINES
#define MASKING
#define SPLINES
#define MY25_MIXING
#ifdef MY25_MIXING
# define N2S2_HORAVG
# define KANTHA_CLAYSON
#endif
#define DJ_GRADPS
#define ANA_SMFLUX
#define ANA_INITIAL
#define ANA_FSOBC
#define ANA_M2OBC
OUTPUT
Nonlinear model elapsed time profile:
Allocation and array initialization .............. 0.513 (61.3738 %)
2D/3D coupling, vertical metrics ................. 0.094 (11.2055 %)
Omega vertical velocity .......................... 0.035 ( 4.2247 %)
Equation of state for seawater ................... 0.095 (11.3984 %)
Total: 0.737 88.2024
All percentages are with respect to total time = 0.836
ROMS/TOMS - Output NetCDF summary for Grid 01:
Analytical header files used:
ROMS/Functionals/ana_initial.h
ROMS/TOMS - Output error ............ exit_flag: 3
ERROR: Abnormal termination: NetCDF OUTPUT.
REASON: NetCDF: Unknown file format
...........................................................................................
..............................................................................................
INPUT EXAMPLE 2
#define UV_ADV
#define UV_COR
#define UV_QDRAG
#define UV_VIS2
#define MIX_S_UV
#define UV_C4ADVECTION
#define MASKING
#define SPLINES
#define MY25_MIXING
#ifdef MY25_MIXING
# define N2S2_HORAVG
# define KANTHA_CLAYSON
#endif
#define DJ_GRADPS
#define ANA_SMFLUX
#define ANA_INITIAL
#define ANA_FSOBC
#define ANA_M2OBC
OUTPUT 2
STEP Day HH:MM:SS KINETIC_ENRG POTEN_ENRG TOTAL_ENRG NET_VOLUME
C => (i,j) Cu Cv C Max Max Speed
0 0 00:00:00 0.000000E+00 0.000000E+00 0.000000E+00 1.403707E+15
(000,000) 0.000000E+00 0.000000E+00 0.000000E+00 0.000000E+00
DEF_HIS - creating history file, Grid 01: sofala_his.nc
WRT_HIS - wrote history fields (Index=1) into time record = 0000001
1 0 00:05:00 0.000000E+00 0.000000E+00 0.000000E+00 1.403707E+15
(000,000) 0.000000E+00 0.000000E+00 0.000000E+00 0.000000E+00
2 0 00:10:00 0.000000E+00 0.000000E+00 0.000000E+00 1.403707E+15
(000,000) 0.000000E+00 0.000000E+00 0.000000E+00 0.000000E+00
-
ShinerM
Re: Minus sediment concentration in river sediment case
Hey Kate,kate wrote:As I said back in March 2011, it depends on how you do your counting. Assuming you are counting in the Matlab/Python/ncview way, the indices of the v-points should match those of the rho-points for the north-to-south rivers and should be one less in j from the rho-points for the south-to-north rivers. In both cases, the i-indices should match those of the rho-points.ShinerM wrote:Thanks so much for your responses.kate wrote:The river guidelines are posted here. Can you provide a drawing of the land mask around your river point and the i,j values?
The files I attached are the river points in the mask_v from my grid file.The river points in the mask_v of the north to south river are (4,3)(5,3)(6,3),so the real river points I should get for the roms running are (3,3)(4,3)(5,3).
So as to the south to north river.The river points in the mask_v are (3,132)(4,132)(5,132),so the real river points I should get for the roms running are (2,132)(3,132)(4,132).Am I right?
When I set these river points in my make_forcing_river.m file,and the result of the roms runnning are strange.the salt value in the river points are all larger than 128.Both rivers have this case.
However,if i set the river points in (3,2)(4,2)(5,2),and (2,133)(3,133)(4,133)(these points are all land points),the salt value seems regular.but the velocity in the river points are all zero.
Waiting for your responses.Thanks.
Are you providing tracer values on inflow? If not, you are facing the dread down-stream advection scheme, known to be unstable.
Thanks for your tips.The problems have already been solved.The roms run well and the salt value becomes normal.Many thanks for your help!
Re: Minus sediment concentration in river sediment case
For Helder, you already had SPLINES defined, but neither it nor SALINITY will do anything without SOLVE3D - that's the important one here. SOLVE3D turns on many options, including the vertical mixing schemes.
Did it say which file it was having trouble with? There might be an error message before the profile output.ERROR: Abnormal termination: NetCDF OUTPUT.
REASON: NetCDF: Unknown file format
-
machaielder
- Posts: 43
- Joined: Wed Mar 18, 2015 3:40 pm
- Location: Universidade Federal Fluminense
Re: Minus sediment concentration in river sediment case
Hello Kate
Thanks for replying.
I think the missing part of error is:
NETCDF_OPEN - unable to open existing NetCDF file:
frc.nc
call from: check_multifile.F, check_file
Elapsed CPU time (seconds):
Thread # 0 CPU: 0.836
Total: 0.836
Nonlinear model elapsed time profile:
Allocation and array initialization .............. 0.513 (61.3738 %)
2D/3D coupling, vertical metrics ................. 0.094 (11.2055 %)
Omega vertical velocity .......................... 0.035 ( 4.2247 %)
Equation of state for seawater ................... 0.095 (11.3984 %)
Total: 0.737 88.2024
All percentages are with respect to total time = 0.836
ROMS/TOMS - Output NetCDF summary for Grid 01:
Analytical header files used:
ROMS/Functionals/ana_initial.h
ROMS/TOMS - Output error ............ exit_flag: 3
I suspect that the model calls for forcing file and does not find...!?
How could I solve it? How do I son turn on SOLVE3D. AS I said I Have salinity and temperature data from WOAD09 and tides from TPX07.1 to force the model. I problem is to include the forcing in the model.
Many Thanks Kate;
REgards, Helder
Thanks for replying.
I think the missing part of error is:
NETCDF_OPEN - unable to open existing NetCDF file:
frc.nc
call from: check_multifile.F, check_file
Elapsed CPU time (seconds):
Thread # 0 CPU: 0.836
Total: 0.836
Nonlinear model elapsed time profile:
Allocation and array initialization .............. 0.513 (61.3738 %)
2D/3D coupling, vertical metrics ................. 0.094 (11.2055 %)
Omega vertical velocity .......................... 0.035 ( 4.2247 %)
Equation of state for seawater ................... 0.095 (11.3984 %)
Total: 0.737 88.2024
All percentages are with respect to total time = 0.836
ROMS/TOMS - Output NetCDF summary for Grid 01:
Analytical header files used:
ROMS/Functionals/ana_initial.h
ROMS/TOMS - Output error ............ exit_flag: 3
I suspect that the model calls for forcing file and does not find...!?
How could I solve it? How do I son turn on SOLVE3D. AS I said I Have salinity and temperature data from WOAD09 and tides from TPX07.1 to force the model. I problem is to include the forcing in the model.
Many Thanks Kate;
REgards, Helder
Re: Minus sediment concentration in river sediment case
When you turn on SOLVE3D, the model suddenly needs a lot more inputs. You have:
but it will also want ANA_STFLUX and ANA_SSFLUX for surface tracer forcing, plus ANA_BTFLUX, ANA_BSFLUX for the bottom. You can look in globaldefs.h to see what conditions turn on the need for FRC_FILE (forcing file), where I see there's a sediment clause too, asking for ANA_SPFLUX and ANA_BPFLUX.
Code: Select all
#define ANA_SMFLUX-
machaielder
- Posts: 43
- Joined: Wed Mar 18, 2015 3:40 pm
- Location: Universidade Federal Fluminense
Re: Minus sediment concentration in river sediment case
Hello Kate
Thank you
Are you meaning this part of globaldefs.h ? if yes, do I need do define all this inputs in my Sofala.h?
#ifdef SOLVE3D
# ifdef BULK_FLUXES
# ifdef ANA_SMFLUX
# undef ANA_SMFLUX
# endif
# ifdef ANA_STFLUX
# undef ANA_STFLUX
# endif
# endif
# if !defined ANA_BTFLUX || \
(!defined AIR_OCEAN && \
!defined BULK_FLUXES && !defined ANA_SMFLUX) || \
(!defined BULK_FLUXES && !defined ANA_STFLUX) || \
( defined BIOLOGY && !defined ANA_SPFLUX) || \
( defined BIOLOGY && !defined ANA_BPFLUX) || \
( defined BULK_FLUXES && !defined LONGWAVE) || \
( defined BULK_FLUXES && !defined ANA_PAIR) || \
( defined BULK_FLUXES && !defined ANA_TAIR) || \
( defined BULK_FLUXES && !defined ANA_HUMIDITY) || \
( defined BULK_FLUXES && !defined ANA_CLOUD) || \
( defined BULK_FLUXES && !defined ANA_RAIN) || \
( defined BULK_FLUXES && !defined ANA_WINDS) || \
( defined BULK_FLUXES && !defined ANA_SRFLUX) || \
( defined LMD_SKPP && !defined ANA_SRFLUX) || \
( defined SALINITY && !defined ANA_SSFLUX) || \
( defined SOLAR_SOURCE && !defined ANA_SRFLUX) || \
( defined SSH_TIDES || defined UV_TIDES) || \
( defined BBL_MODEL && (!defined ANA_WWAVE && \
!defined WAVES_OCEAN)) || \
( defined SEDIMENT && !defined ANA_SPFLUX) || \
( defined SEDIMENT && !defined ANA_BPFLUX) || \
( defined WAVE_DATA && (!defined ANA_WWAVE && \
!defined WAVES_OCEAN))
# define FRC_FILE
# endif
#else
# if !defined ANA_SMFLUX || \
( defined SSH_TIDES || defined UV_TIDES)
# define FRC_FILE
# endif
#endif
Thank you
Are you meaning this part of globaldefs.h ? if yes, do I need do define all this inputs in my Sofala.h?
#ifdef SOLVE3D
# ifdef BULK_FLUXES
# ifdef ANA_SMFLUX
# undef ANA_SMFLUX
# endif
# ifdef ANA_STFLUX
# undef ANA_STFLUX
# endif
# endif
# if !defined ANA_BTFLUX || \
(!defined AIR_OCEAN && \
!defined BULK_FLUXES && !defined ANA_SMFLUX) || \
(!defined BULK_FLUXES && !defined ANA_STFLUX) || \
( defined BIOLOGY && !defined ANA_SPFLUX) || \
( defined BIOLOGY && !defined ANA_BPFLUX) || \
( defined BULK_FLUXES && !defined LONGWAVE) || \
( defined BULK_FLUXES && !defined ANA_PAIR) || \
( defined BULK_FLUXES && !defined ANA_TAIR) || \
( defined BULK_FLUXES && !defined ANA_HUMIDITY) || \
( defined BULK_FLUXES && !defined ANA_CLOUD) || \
( defined BULK_FLUXES && !defined ANA_RAIN) || \
( defined BULK_FLUXES && !defined ANA_WINDS) || \
( defined BULK_FLUXES && !defined ANA_SRFLUX) || \
( defined LMD_SKPP && !defined ANA_SRFLUX) || \
( defined SALINITY && !defined ANA_SSFLUX) || \
( defined SOLAR_SOURCE && !defined ANA_SRFLUX) || \
( defined SSH_TIDES || defined UV_TIDES) || \
( defined BBL_MODEL && (!defined ANA_WWAVE && \
!defined WAVES_OCEAN)) || \
( defined SEDIMENT && !defined ANA_SPFLUX) || \
( defined SEDIMENT && !defined ANA_BPFLUX) || \
( defined WAVE_DATA && (!defined ANA_WWAVE && \
!defined WAVES_OCEAN))
# define FRC_FILE
# endif
#else
# if !defined ANA_SMFLUX || \
( defined SSH_TIDES || defined UV_TIDES)
# define FRC_FILE
# endif
#endif
Re: Minus sediment concentration in river sediment case
No, no, no, this does not go in your case.h file, it stays in globaldefs.h. globaldefs.h is parsed after your file and modifies what gets defined. For you, FRC_FILE was defined, but you didn't provide a forcing file. As soon as you turn on SOLVE3D, the model needs surface and bottom tracer forcing, whether through ANA_xxx or a forcing file. You decide which and tell the model with your ANA_xxx choices.
-
machaielder
- Posts: 43
- Joined: Wed Mar 18, 2015 3:40 pm
- Location: Universidade Federal Fluminense
Re: Minus sediment concentration in river sediment case
Hello Kate, thanks
I made the following sets the model runs..but gives me a constant value of salt and temp...and didn't bring sediment.
what should be the cause of no changes in salt and temp. if it is caused by using analytical forcing how should I do to change it.
#define SALINITY
#define SOLVE3D
#define SPLINES
#define ANA_INITIAL
#define ANA_SMFLUX
#define ANA_STFLUX
#define ANA_SSFLUX
#define ANA_BTFLUX
#define ANA_BSFLUX
GRDNAME == sofala_grd.nc
ININAME == ocean_ini.nc
ITLNAME == ocean_itl.nc
IRPNAME == ocean_irp.nc
IADNAME == ocean_iad.nc
FWDNAME == ocean_fwd.nc
ADSNAME == ocean_ads.nc
I made the following sets the model runs..but gives me a constant value of salt and temp...and didn't bring sediment.
what should be the cause of no changes in salt and temp. if it is caused by using analytical forcing how should I do to change it.
#define SALINITY
#define SOLVE3D
#define SPLINES
#define ANA_INITIAL
#define ANA_SMFLUX
#define ANA_STFLUX
#define ANA_SSFLUX
#define ANA_BTFLUX
#define ANA_BSFLUX
GRDNAME == sofala_grd.nc
ININAME == ocean_ini.nc
ITLNAME == ocean_itl.nc
IRPNAME == ocean_irp.nc
IADNAME == ocean_iad.nc
FWDNAME == ocean_fwd.nc
ADSNAME == ocean_ads.nc
Re: Minus sediment concentration in river sediment case
Well, what are you expecting to have happen? What is the set up? What is the forcing? How should it evolve?
-
machaielder
- Posts: 43
- Joined: Wed Mar 18, 2015 3:40 pm
- Location: Universidade Federal Fluminense
Re: Minus sediment concentration in river sediment case
Hello Kate
perhaps I need to improve my understand in relation to forcing;
I am forcing it with SST, SSS, surface v momentum stress, u momentum stress; (FRCNAME ==)
I expected that to have time varying outputs.
another issue
I would like to locate the River and define sediments while I work out the forcing and initial conditions.
so I would like to know how to set the position of a river in the model.
Regards
Helder
perhaps I need to improve my understand in relation to forcing;
I am forcing it with SST, SSS, surface v momentum stress, u momentum stress; (FRCNAME ==)
I expected that to have time varying outputs.
another issue
I would like to locate the River and define sediments while I work out the forcing and initial conditions.
so I would like to know how to set the position of a river in the model.
Regards
Helder
Re: Minus sediment concentration in river sediment case
Search this forum and the wiki for river sources and you'll come up with some answers.
Forcing with SSS and SST is one way to get started, maybe for a simplified experiment. For realistic applications, I always force with BULK_FLUXES (the CCSM variant), winds and other atmospheric fields from model reanalyses. You really need to think about the purpose of your runs and what you want to learn from them.
For future questions, perhaps start a new thread and state your goals more clearly - best hindcast? Learn something about sediments in an idealized case? Prepare for future simulations using future atmospheric fields? What time scales?
Forcing with SSS and SST is one way to get started, maybe for a simplified experiment. For realistic applications, I always force with BULK_FLUXES (the CCSM variant), winds and other atmospheric fields from model reanalyses. You really need to think about the purpose of your runs and what you want to learn from them.
For future questions, perhaps start a new thread and state your goals more clearly - best hindcast? Learn something about sediments in an idealized case? Prepare for future simulations using future atmospheric fields? What time scales?