Hello!
While trying to execute my ROMS model which has a nested grid in it, I get the following error:
sbasak08@DESKTOP-UOIVLQT:~/roms/Projects/philippines$ ./romsG <External/roms_philippines.in
--------------------------------------------------------------------------------
Model Input Parameters: ROMS/TOMS version 3.9
Tuesday - June 1, 2021 - 10:36:31 AM
--------------------------------------------------------------------------------
LOAD_LBC - incorrect continuation symbol in line:
Clo Clo Clo Clo \ ! salinity,Grid 1
number of nested grid values exceeded.
Found Error: 02 Line: 3407 Source: ROMS/Utility/read_phypar.F
Found Error: 02 Line: 191 Source: ROMS/Utility/inp_par.F
Found Error: 02 Line: 115 Source: ROMS/Drivers/nl_ocean.h, ROMS_initialize
Elapsed wall CPU time for each process (seconds):
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
Dynamic and Automatic memory (MB) usage for Grid 01: 298x148x15 tiling: 1x1
tile Dynamic Automatic USAGE
0 1.80 11.52 13.32
SUM 1.80 11.52 13.32
Dynamic and Automatic memory (MB) usage for Grid 02: 298x148x15 tiling: 1x1
tile Dynamic Automatic USAGE
0 0.00 11.52 11.52
SUM 0.00 11.52 11.52
TOTAL 1.80 23.04 24.84
<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<
ROMS/TOMS - Output NetCDF summary for Grid 01:
close_io.f90:151: runtime error: signed integer overflow: -4702111234474983746 * -4702111234474983746 cannot be represented in type 'integer(kind=8)'
close_io.f90:151: runtime error: signed integer overflow: -4815586015397456958 * -4702111234474983746 cannot be represented in type 'integer(kind=8)'
close_io.f90:151: runtime error: load of misaligned address 0xb44df19f5718e4ba for type 'integer(kind=4)', which requires 4 byte alignment
0xb44df19f5718e4ba: note: pointer points here
<memory cannot be printed>
Program received signal SIGSEGV: Segmentation fault - invalid memory reference.
Backtrace for this error:
#0 0x7f3dff060d01 in ???
#1 0x7f3dff05fed5 in ???
#2 0x7f3dfe39620f in ???
#3 0x7f3e05c79709 in close_out_
at /home/sbasak08/roms/Projects/philippines/Build_romsG/close_io.f90:151
#4 0x7f3e03bc4d4b in __ocean_control_mod_MOD_roms_finalize
at /home/sbasak08/roms/Projects/philippines/Build_romsG/ocean_control.f90:265
#5 0x7f3e03bc2391 in ocean
at /home/sbasak08/roms/Projects/philippines/Build_romsG/master.f90:93
#6 0x7f3e03bc24db in main
at /home/sbasak08/roms/Projects/philippines/Build_romsG/master.f90:50
Segmentation fault (core dumped)
sbasak08@DESKTOP-UOIVLQT:~/roms/Projects/philippines$
I am not sure what might be causing this "LOAD_LBC - incorrect continuation symbol in line: Clo Clo Clo Clo \ ! salinity, Grid number of nested grid values exceeded" error. I am using 1 child grid with 1 parent grid. According to
https://www.myroms.org/wiki/Lake_Jersey ... Example_AB , where they've used 1 nested grid with a parent they too have used:
Ngrids = 2
NestLayers = 2
GridsInLayer = 1 1
LBC(isFsur) == Clo Clo Clo Clo \ ! free-surface, Grid 1
Nes Nes Nes Nes ! free-surface, Grid 2
LBC(isUbar) == Clo Clo Clo Clo \ ! 2D U-momentum, Grid 1
Nes Nes Nes Nes ! 2D U-momentum, Grid 2
LBC(isVbar) == Clo Clo Clo Clo \ ! 2D V-momentum, Grid 1
Nes Nes Nes Nes ! 2D V-momentum, Grid 2
LBC(isUvel) == Clo Clo Clo Clo \ ! 3D U-momentum, Grid 1
Nes Nes Nes Nes ! 3D U-momentum, Grid 2
LBC(isVvel) == Clo Clo Clo Clo \ ! 3D V-momentum, Grid 1
Nes Nes Nes Nes ! 3D V-momentum, Grid 2
LBC(isMtke) == Clo Clo Clo Clo \ ! mixing TKE, Grid 1
Nes Nes Nes Nes ! mixing TKE, Grid 2
LBC(isTvar) == Clo Clo Clo Clo \ ! temperature, Grid 1
Clo Clo Clo Clo \ ! salinity, Grid 1
Nes Nes Nes Nes \ ! temperature, Grid 2
Even I am using the same script structure as shown above.
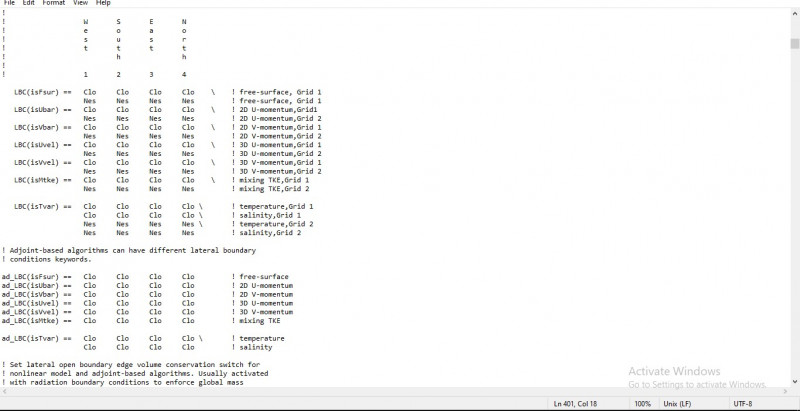
- part of my roms_philippines.in file
Is that because of the grid file I am using? Or the nested one/contact one? I can see that the h min and max is changing to NaN, is that a possible reason for this error? I created my child grid using Matlab script of coarse2fine.m from the ROMS Matlab repository and I got the file
with an output in the command window like this:
>> Imin = 153;
>> Imax = 202;
>> Jmin = 43;
>> Jmax = 74;
>> C = coarse2fine ('philippines_parent.nc', 'philippines_child_ceb.nc', 3, Imin, Imax, Jmin, Jmax);
Interpolating from coarse to fine ...
Computing grid spacing: great circle distances
Number of points: Coarse = 300 x 150, fine = 149 x 95
Requested global attribute "history" not found in: philippines_child_ceb.nc
Writing finer grid variables into: philippines_child_ceb.nc
Wrote f Min= 2.41572e-05 Max= 3.04438e-05
Wrote h Min= NaN Max= NaN
Wrote pm Min= 5.29309e-04 Max= 5.29309e-04
Wrote pn Min= 3.02453e-04 Max= 3.04994e-04
Wrote dmde Min=-2.38549e-06 Max= 2.38549e-04
If the error's due to the build file as the read_phypar.f is present in the build_romsG folder, should I be building it again to solve the issue or the cpps defs are incorrectly written in my header file? My header file looks like this:
/*
** svn $Id: lake_jersey.h 995 2020-01-10 04:01:28Z arango $
*******************************************************************************
** Copyright (c) 2002-2020 The ROMS/TOMS Group **
** Licensed under a MIT/X style license **
** See License_ROMS.txt **
*******************************************************************************
**
** Options for Lake Jersey forced with wind.
**
** Application flag: Philippines
**
** Input scripts: roms_philippines.in
**
*/
#define UV_ADV
#define UV_COR
#define UV_QDRAG
#define UV_VIS4
#define MIX_S_UV
#define DJ_GRADPS
#define SPLINES_VDIFF
#define SPLINES_VVISC
#define BULK_FLUXES
#define SOLVE3D
#define BODYFORCE
#define NESTING
#define ONE_WAY
#define ANA_SMFLUX
#define ANA_STFLUX
#define ANA_INITIAL
#define ANA_SSFLUX
#define ANA_DRAG
#define ANA_BSFLUX
#define ANA_BTFLUX
#define MASKING
What might be the reason behind this error and is there any way to resolve it?
Any help is deeply appreciated by me.
Kind regards
-Neel