The latest version of the
Many thanks to Andy Moore for his great help in the development and testing of all 4D-Var and (4D-Var)T data assimilation algorithms.
What is New:
- The test
 svn repository was updated to include several idealized and realistic ROMS applications. Notice that the repository includes several
svn repository was updated to include several idealized and realistic ROMS applications. Notice that the repository includes several  sub-directories. Each sub-directory contains the necessary header file (my_application.h), input scripts (ocean.in, and others), build scripts (build.bash, build.sh), and, if applicable, configuration scripts (job_*.in). These test cases show the versatility of the build script and how users can play with different combinations of C-preprocessing options, ROMS source code, parallel/serial applications, and various MPI libraries (mpich, mpich2, OpenMPI). Notice that each application header file is very clean (no #undef directives) and the important CPP options are actually defined in the build script, for example in the default upwelling test case we have:
Notice that it is much easier to comment options in the build script that using the define/undef directive in the header file.
sub-directories. Each sub-directory contains the necessary header file (my_application.h), input scripts (ocean.in, and others), build scripts (build.bash, build.sh), and, if applicable, configuration scripts (job_*.in). These test cases show the versatility of the build script and how users can play with different combinations of C-preprocessing options, ROMS source code, parallel/serial applications, and various MPI libraries (mpich, mpich2, OpenMPI). Notice that each application header file is very clean (no #undef directives) and the important CPP options are actually defined in the build script, for example in the default upwelling test case we have:
Notice that it is much easier to comment options in the build script that using the define/undef directive in the header file.Code: Select all
export MY_CPP_FLAGS="-DAVERAGES" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DDIAGNOSTICS_TS" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DDIAGNOSTICS_UV" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DSTATIONS" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DFLOATS" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DFLOAT_VWALK" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DDEBUGGING" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DOUT_DOUBLE" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DEW_PERIODIC" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DUV_VIS2" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DUV_VIS4" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DMIX_S_UV" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DMIX_GEO_UV" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DUV_LOGDRAG" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DUV_LDRAG" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DUV_QDRAG" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DUV_DRAG_GRID" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DANA_DRAG" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DTS_U3HADVECTION" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DTS_C4VADVECTION" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DTS_MPDATA" export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DBIO_FENNEL" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DECOSIM" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DNEMURO" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DNPZD_FRANKS" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DNPZD_IRON" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DNPZD_POWELL" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DGLS_MIXING" #export MY_CPP_FLAGS="${MY_CPP_FLAGS} -DMY25_MIXING" - To checkout the test cases repository use:
Code: Select all
svn checkout --username joe_roms https://www.myroms.org/svn/src/test my_test - We added test cases for the 4D-Var and (4D-Var)T data assimilation algorithms using California Current System, 1/3 resolution, WC13 application. You will find a directory for each 4D-Var algorithm:
We provided a lot of documentation about how to use these algorithms in
Code: Select all
/WC13 Main California Current System 4D-Var applications /ARRAY_MODES Stabilized representer matrix array modes and clipping /Data Input data directory /Functionals Analytical expression header file /I4DVAR Primal form of incremental, strong constraint 4D-Var, I4D-Var /I4DVAR_impact I4D-Var observation impact /Normalization 4D-Var error covariance normalization coefficients /plotting 4D-Var plotting scripts (Matlab and ROMS plotting package) /PSAS Dual form of 4D-Var, Physical-space Statistical Analysis System, 4D-PSAS /PSAS_impact 4D-PSAS observation impact /PSAS_sensitivity 4D-PSAS observation sensitivity (adjoint of 4D-PSAS) /R4DVAR Dual form of 4D-Var, indirect representer method, R4D-Var /R4DVAR_impact R4D-Var observation impact /R4DVAR_sensitivity R4D-Var observation sensitivity (adjoint of R4D-Var) WikiROMS with several 4D-Var tutorials. The tutorials are part of the ROMS data assimilation workshop that Andy Moore and I offered in July 2010 at the University of California, Santa Cruz.
WikiROMS with several 4D-Var tutorials. The tutorials are part of the ROMS data assimilation workshop that Andy Moore and I offered in July 2010 at the University of California, Santa Cruz. - Added test cases for the adjoint-based Generalized Stability Theory (GST) algorithms using the idealized double gyre example. You will find a directory for each GST algorithm:
Code: Select all
/double_gyre Main double gyre test case /Data Input NetCDF file /Forward Nonlinear forward trajectory /AFTE Adjoint Finite Time Eigenmodes /FTE Finite time Eigenmodes /FSV Forcing Singular Vectors /OP Optimal Perturbations /SOsemi Stochastic Optimals, seminorn - Added grid, initial conditions, and forcing NetCDF files for the BIO_TOY test case. It includes initial conditions for BIO_FENNEL, ECOSIM, NEMURO, and all NPZD models (NPZD_FRANKS, NPZD_IRON, and NPZD_POWELL). All the NetCDF files are now under svn control and located in the bio_toy/Data sub-directory. The grid is a 5x5 double periodic domain sampled from the North Eastern North Atlantic (NENA) application. There is an initial conditions file corresponding to each ecosystem model. The fields are horizontally uniform rendering ROMS into 1D vertical model. Currently, we have the following files:
The forcing data is from a typical year (days 0.5 to 364.5) ECMWF 40-year re-analysis. The time coordinate has the cycle_length attribute with a 365 value. Therefore, this application can be run for several years with a perpetual annual forcing cycle.
Code: Select all
bio_toy/Data/bio_toy_grd.nc grid bio_toy/Data/bio_toy_frc.nc forcing bio_toy/Data/bio_toy_ini_fennel.nc BIO_FENNEL bio_toy/Data/bio_toy_ini_ecosim.nc ECOSIM bio_toy/Data/bio_toy_ini_nemuro.nc NEMURO bio_toy/Data/bio_toy_ini_npzd.nc any NPZD model - The build script (ROMS/Bin/build.bash and ROMS/Bin/build.sh) was improved and updated. Most of the user tunable parameters are close to each other and better documented:
Notice the new local variable which_MPI to specify which MPI library to use during compilation. You need choose only one and comment the other values. The value of local variable USE_MY_LIBS is commented out by default. If you want to use this nice capability, you need to change the paths of the required libraries in your particular computer. Recall that you may want to have different copies of the build script for a particular computer.
Code: Select all
# Other user defined environmental variables. See the ROMS makefile for # details on other options the user might want to set here. Be sure to # leave the switches meant to be off set to an empty string or commented # out. Any string value (including off) will evaluate to TRUE in # conditional if-statements. export USE_MPI=on # distributed-memory parallelism export USE_MPIF90=on # compile with mpif90 script #export which_MPI=mpich # compile with MPICH library #export which_MPI=mpich2 # compile with MPICH2 library export which_MPI=openmpi # compile with OpenMPI library #export USE_OpenMP=on # shared-memory parallelism export FORT=ifort #export FORT=gfortran #export FORT=pgi #export USE_DEBUG=on # use Fortran debugging flags export USE_LARGE=on # activate 64-bit compilation #export USE_NETCDF4=on # compile with NetCDF-4 library #export USE_PARALLEL_IO=on # Parallel I/O with Netcdf-4/HDF5 #export USE_MY_LIBS=on # use my library paths below - Now we are using the NetCDF-4 configuration script nc-config which is available since NetCDF version 4.0.1. This script reports all the required libraries according to the NetCDF-4 installation options (openDAP, NetCDF4/HDF5 file format). For Example, the logic for the ifort compiler in build.bash is now:
Again, all the above logic is processed when the environmental variable USE_MY_LIBS is not empty and has any non-blank value. This change required an update to all the make configuration (*.mk) files in the Compilers directory. This cleaned the logic nicely. Notice that nc-config is not available for NetCDF 3.x. Many thanks to Mark Hadfield for
Code: Select all
if [ -n "${USE_MY_LIBS:+1}" ]; then case "$FORT" in ifort ) export ESMF_OS=Linux export ESMF_COMPILER=ifort export ESMF_BOPT=O export ESMF_ABI=64 export ESMF_COMM=mpich export ESMF_SITE=default export ARPACK_LIBDIR=/opt/intelsoft/serial/ARPACK if [ -n "${USE_MPI:+1}" ]; then if [ "${which_MPI}" = "mpich" ]; then export ESMF_DIR=/opt/intelsoft/mpich/esmf export MCT_INCDIR=/opt/intelsoft/mpich/mct/include export MCT_LIBDIR=/opt/intelsoft/mpich/mct/lib export PARPACK_LIBDIR=/opt/intelsoft/mpich/PARPACK elif [ "${which_MPI}" = "mpich2" ]; then export ESMF_DIR=/opt/intelsoft/mpich2/esmf export MCT_INCDIR=/opt/intelsoft/mpich2/mct/include export MCT_LIBDIR=/opt/intelsoft/mpich2/mct/lib export PARPACK_LIBDIR=/opt/intelsoft/mpich2/PARPACK elif [ "${which_MPI}" = "openmpi" ]; then export ESMF_DIR=/opt/intelsoft/openmpi/esmf export MCT_INCDIR=/opt/intelsoft/openmpi/mct/include export MCT_LIBDIR=/opt/intelsoft/openmpi/mct/lib export PARPACK_LIBDIR=/opt/intelsoft/openmpi/PARPACK fi fi if [ -n "${USE_NETCDF4:+1}" ]; then if [ -n "${USE_PARALLEL_IO:+1}" ] && [ -n "${USE_MPI:+1}" ]; then if [ "${which_MPI}" = "mpich" ]; then export NC_CONFIG=/opt/intelsoft/mpich/netcdf4/bin/nc-config export NETCDF_INCDIR=/opt/intelsoft/mpich/netcdf4/include elif [ "${which_MPI}" = "mpich2" ]; then export NC_CONFIG=/opt/intelsoft/mpich2/netcdf4/bin/nc-config export NETCDF_INCDIR=/opt/intelsoft/mpich2/netcdf4/include elif [ "${which_MPI}" = "openmpi" ]; then export NC_CONFIG=/opt/intelsoft/openmpi/netcdf4/bin/nc-config export NETCDF_INCDIR=/opt/intelsoft/openmpi/netcdf4/include fi else export NC_CONFIG=/opt/intelsoft/serial/netcdf4/bin/nc-config export NETCDF_INCDIR=/opt/intelsoft/serial/netcdf4/include fi else export NETCDF_INCDIR=/opt/intelsoft/serial/netcdf3/include export NETCDF_LIBDIR=/opt/intelsoft/serial/netcdf3/lib fi ;; ... suggesting this change.
suggesting this change.
We highly recommend using the build script when compiling and linking ROMS. Advanced users that run on various computer architecture may want to have their own Compilers sub-directory somewhere else. I have my customized make configuration files in $HOME/Compilers. So I tell the build script to use those configuration files instead:Code: Select all
#export COMPILERS=${MY_ROMS_SRC}/Compilers export COMPILERS=${HOME}/Compilers - The time-averaged, diagnostic terms for the momentum (DIAGNOSTICS_UV) and tracer (DIAGNOSTICS_TS) equations were expanded to include components in the ξ- and η-directions for horizontal advection and horizontal viscosity/diffusion. For technical information on implementation check following
 trac ticket.
trac ticket.
- The momentum advection output variables are:
Code: Select all
ubar_hadv, vbar_hadv 2D momentum horizontal advection ubar_xadv, vbar_xadv 2D momentum horizontal ξ-advection ubar_yadv, vbar_yadv 2D momentum horizontal η-advection u_hadv, v_hadv 3D momentum horizontal advection u_xadv, v_xadv 3D momentum horizontal ξ-advection u_yadv, v_yadv 3D momentum horizontal η-advection u_vadv, v_vadv 3D momentum vertical advection - The momentum viscosity (stress tensor) output variables are:
Code: Select all
ubar_hvisc, vbar_hvisc 2D momentum horizontal viscosity ubar_xvisc, vbar_xvisc 2D momentum horizontal ξ-viscosity ubar_yvisc, vbar_yvisc 2D momentum horizontal η-viscosity u_hvisc, v_hvisc 3D momentum horizontal viscosity u_xvisc, v_xvisc 3D momentum horizontal ξ-viscosity u_yvisc, v_yvisc 3D momentum horizontal η-viscosity u_vvisc, v_vvisc 3D momentum vertical viscosity - The tracer advection output variables are: (Tname is typically, temp, salt, ...)
Code: Select all
Tname_hadv tracer horizontal advection Tname_xadv tracer horizontal ξ-advection Tname_yadv tracer horizontal η-advection Tname_vadv tracer vertical advection - The tracer diffusion output variables are: (Tname is typically, temp, salt, ...)
Code: Select all
Tname_hdiff tracer horizontal diffusion Tname_xdiff tracer horizontal ξ-diffusion Tname_ydiff tracer horizontal η-diffusion Tname_sdiff tracer horizontal s-diffusion (rotated tensor) Tname_vdiff tracer vertical diffusion - Vorticity diagnostics output variables (new fields computed in new routine vorticity.F):
Code: Select all
rvorticity_bar 2D relative vorticity pvorticity_bar 2D potential vorticity rvorticity 3D relative vorticity pvorticity 3D potential vorticity
- The momentum advection output variables are:
- The logic for writing time-averaged fields (AVERAGES) and diagnostic terms (DIAGNOSTICS_TS, DIAGNOSTICS_UV) into respective output NetCDF files was redesigned to provide more flexibility and facilitate expansion in the future. A new set of logical switches (T/F) Aout were added to control the time-averaged fields written into the output AVGNAME NetCDF file. These switches are set in the standard input script, ocean.in:
The strategy here is to provide the user with switches to control which variables are written into the AVGNAME output file. The time-averages require global arrays to hold the accumulated sums. This increases the memory requirements of the model. This update only allocates those fields that are requested. Otherwise, the pointer for the array in the AVERAGE(ng)%avg... structure remains unassociated and does not use memory. For example, in mod_average.F we now have:
Code: Select all
! Logical switches (TRUE/FALSE) to activate writing of time-averaged ! fields into AVERAGE output file. Aout(idUvel) == T ! u 3D U-velocity Aout(idVvel) == T ! v 3D V-velocity Aout(idWvel) == T ! w 3D W-velocity Aout(idOvel) == T ! omega omega vertical velocity Aout(idUbar) == T ! ubar 2D U-velocity Aout(idVbar) == T ! vbar 2D V-velocity Aout(idFsur) == T ! zeta free-surface Aout(idTvar) == T T ! temp, salt temperature and salinity Aout(idUsms) == F ! sustr surface U-stress Aout(idVsms) == F ! svstr surface V-stress Aout(idUbms) == F ! bustr bottom U-stress Aout(idVbms) == F ! bvstr bottom V-stress Aout(idW2xx) == F ! Sxx_bar 2D radiation stress, Sxx component Aout(idW2xy) == F ! Sxy_bar 2D radiation stress, Sxy component Aout(idW2yy) == F ! Syy_bar 2D radiation stress, Syy component Aout(idU2rs) == F ! Ubar_Rstress 2D radiation U-stress Aout(idV2rs) == F ! Vbar_Rstress 2D radiation V-stress Aout(idU2Sd) == F ! ubar_stokes 2D U-Stokes velocity Aout(idV2Sd) == F ! vbar_stokes 2D V-Stokes velocity Aout(idW3xx) == F ! Sxx 3D radiation stress, Sxx component Aout(idW3xy) == F ! Sxy 3D radiation stress, Sxy component Aout(idW3yy) == F ! Syy 3D radiation stress, Syy component Aout(idW3zx) == F ! Szx 3D radiation stress, Szx component Aout(idW3zy) == F ! Szy 3D radiation stress, Szy component Aout(idU3rs) == F ! u_Rstress 3D U-radiation stress Aout(idV3rs) == F ! v_Rstress 3D V-radiation stress Aout(idU3Sd) == F ! u_stokes 3D U-Stokes velocity Aout(idV3Sd) == F ! v_stokes 3D V-Stokes velocity Aout(idPair) == F ! Pair surface air pressure Aout(idUair) == F ! Uair surface U-wind component Aout(idVair) == F ! Vair surface V-wind component Aout(idTsur) == F F ! shflux, ssflux surface net heat and salt flux Aout(idLhea) == F ! latent latent heat flux Aout(idShea) == F ! sensible sensible heat flux Aout(idLrad) == F ! lwrad longwave radiation flux Aout(idSrad) == F ! swrad shortwave radiation flux Aout(idevap) == F ! evaporation evaporation rate Aout(idrain) == F ! rain precipitation rate Aout(idDano) == F ! rho density anomaly Aout(idVvis) == F ! AKv vertical viscosity Aout(idTdif) == F ! AKt vertical T-diffusion Aout(idSdif) == F ! AKs vertical Salinity diffusion Aout(idHsbl) == F ! Hsbl depth of surface boundary layer Aout(idHbbl) == F ! Hbbl depth of bottom boundary layer Aout(idHUav) == F ! Huon u-volume flux, Huon Aout(idHVav) == F ! Hvom v-volume flux, Hvom Aout(idUUav) == F ! uu quadratic <u*u> term Aout(idUVav) == F ! uv quadratic <u*v> term Aout(idVVav) == F ! vv quadratic <v*v> term Aout(idU2av) == F ! ubar2 quadratic <ubar*ubar> term Aout(idV2av) == F ! vbar2 quadratic <vbar*vbar> term Aout(idZZav) == F ! zeta2 quadratic <zeta*zeta> term Aout(idTTav) == F F ! temp2, salt2 quadratic <t*t> T/S terms Aout(idUTav) == F F ! utemp, usalt quadratic <u*t> T/S terms Aout(idVTav) == F F ! vtemp, vsalt quadratic <v*t> T/S terms Aout(iHUTav) == F F ! Huontemp, ... T/S volume flux, <Huon*t> Aout(iHVTav) == F F ! Hvomtemp, ... T/S volume flux, <Hvom*t> ! Logical switches (TRUE/FALSE) to activate writing of extra inert passive ! tracers other than biological and sediment tracers into the AVERAGE file. Aout(inert) == T ! dye_01, ... inert passive tracersSince we control the time-averaged fields with switches, there is no longer a need for the following C-preprocessing options: AVERAGES_AKV, AVERAGES_AKS, AVERAGES_BEDLOAD, AVERAGES_FLUXES, AVERAGES_NEARSHORE, AVERAGES_QUADRATIC. These options are eliminated. The only option that is required to write any of these fields is AVERAGES. This is nice because we do not have to recompile if new fields are needed later.Code: Select all
IF (Aout(idFsur,ng)) THEN allocate ( AVERAGE(ng) % avgzeta(LBi:UBi,LBj:UBj) ) END IF - The routine set_avg_tile was completely redesigned to use these control swithes. Notice that arrays are no longer passed as arguments. This facilitates adding new fields in the future.
Code: Select all
CALL set_avg_tile (ng, tile, & & LBi, UBi, LBj, UBj, & & IminS, ImaxS, JminS, JmaxS, & # ifdef SOLVE3D & NOUT, & # endif & KOUT) ... IF (Aout(idFsur,ng)) THEN DO j=JstrR,JendR DO i=IstrR,IendR AVERAGE(ng)%avgzeta(i,j)=AVERAGE(ng)%avgzeta(i,j)+ & & OCEAN(ng)%zeta(i,j,Kout) END DO END DO END IF - We still need the option AVERAGES_DETIDE to process time-averaged, detided fields. Notice that we also control the processing of these fields with the Aout switches. Also, I added the capability to detide temperature and salinity since we have better control of the memory requirements.
Code: Select all
Aout(idu3dD) == F ! u_detided detided 3D U-velocity Aout(idv3dD) == F ! v_detided detided 3D V-velocity Aout(idu2dD) == F ! ubar_detided detided 2D U-velocity Aout(idv2dD) == F ! vbar_detided detided 2D V-velocity Aout(idFsuD) == F ! zeta_detided detided free-surface Aout(idTrcD) == F F ! temp_detided, ... detided temperature and salinity - The time-averaged vorticity fields were moved from DIANAME to AVGNAME. This is a better place for such fields:
Code: Select all
Aout(id2dRV) == F ! pvorticity_bar 2D relative vorticity Aout(id3dRV) == F ! pvorticity 3D relative vorticity Aout(id2dPV) == F ! rvorticity_bar 2D potential vorticity Aout(id3dPV) == F ! rvorticity 3D potential vorticity - A new set of logical switches (T/F) Dout were added to control the time-averaged diagnostic terms written into the output DIANAME NetCDF file. These switches are set in standard input script, ocean.in:
Similar Aout and Dout switches are added to all the biology and sediment models input scripts. For example, in bio_Fennel.in now we have:
Code: Select all
! Logical switches (TRUE/FALSE) to activate writing of time-averaged, ! 2D momentum (ubar,vbar) diagnostic terms into DIAGNOSTIC output file. Dout(M2rate) == T ! ubar_accel, ... acceleration Dout(M2pgrd) == T ! ubar_prsgrd, ... pressure gradient Dout(M2fcor) == T ! ubar_cor, ... Coriolis force Dout(M2hadv) == T ! ubar_hadv, ... horizontal total advection Dout(M2xadv) == T ! ubar_xadv, ... horizontal XI-advection Dout(M2yadv) == T ! ubar_yadv, ... horizontal ETA-advection Dout(M2hrad) == T ! ubar_hrad, ... horizontal total radiation stress Dout(M2hvis) == T ! ubar_hvisc, ... horizontal total viscosity Dout(M2xvis) == T ! ubar_xvisc, ... horizontal XI-viscosity Dout(M2yvis) == T ! ubar_yvisc, ... horizontal ETA-viscosity Dout(M2sstr) == T ! ubar_sstr, ... surface stress Dout(M2bstr) == T ! ubar_bstr, ... bottom stress ! Logical switches (TRUE/FALSE) to activate writing of time-averaged, ! 3D momentum (u,v) diagnostic terms into DIAGNOSTIC output file. Dout(M3rate) == T ! u_accel, ... acceleration Dout(M3pgrd) == T ! u_prsgrd, ... pressure gradient Dout(M3fcor) == T ! u_cor, ... Coriolis force Dout(M3hadv) == T ! u_hadv, ... horizontal total advection Dout(M3xadv) == T ! u_xadv, ... horizontal XI-advection Dout(M3yadv) == T ! u_yadv, ... horizontal ETA-advection Dout(M3vadv) == T ! u_vadv, ... vertical advection Dout(M3hrad) == T ! u_hrad, ... horizontal total radiation stress Dout(M3vrad) == T ! u_vrad, ... vertical radiation stress Dout(M3hvis) == T ! u_hvisc, ... horizontal total viscosity Dout(M3xvis) == T ! u_xvisc, ... horizontal XI-viscosity Dout(M3yvis) == T ! u_yvisc, ... horizontal ETA-viscosity Dout(M3vvis) == T ! u_vvisc, ... vertical viscosity ! Logical switches (TRUE/FALSE) to activate writing of time-averaged, ! active (temperature and salinity) and passive (inert) tracer diagnostic ! terms into DIAGNOSTIC output file: [1:NAT+NPT,Ngrids]. Dout(iTrate) == T T ! temp_rate, ... time rate of change Dout(iThadv) == T T ! temp_hadv, ... horizontal total advection Dout(iTxadv) == T T ! temp_xadv, ... horizontal XI-advection Dout(iTyadv) == T T ! temp_yadv, ... horizontal ETA-advection Dout(iTvadv) == T T ! temp_vadv, ... vertical advection Dout(iThdif) == T T ! temp_hdiff, ... horizontal total diffusion Dout(iTxdif) == T T ! temp_xdiff, ... horizontal XI-diffusion Dout(iTydif) == T T ! temp_ydiff, ... horizontal ETA-diffusion Dout(iTsdif) == T T ! temp_sdiff, ... horizontal S-diffusion Dout(iTvdif) == T T ! temp_vdiff, ... vertical diffusionNotice that diagnostics activated with DIGNOSTICS_BIO in Fennel's model are also controlled with the Dout instead of the Hout switches above. They need to be scaled before writing in wrt_diag.F:Code: Select all
! Logical switches (TRUE/FALSE) to activate writing of time-averaged fields ! into AVERAGE output file: [1:NBT,Ngrids]. Aout(idTvar) == 12*T ! biological tracer ! Logical switches (TRUE/FALSE) to activate writing of time-averaged, ! biological tracer diagnostic terms into DIAGNOSTIC output file: ! [1:NBT,Ngrids]. Dout(iTrate) == 12*T ! time rate of change Dout(iThadv) == 12*T ! horizontal total advection Dout(iTxadv) == 12*T ! horizontal XI-advection Dout(iTyadv) == 12*T ! horizontal ETA-advection Dout(iTvadv) == 12*T ! vertival advection Dout(iThdif) == 12*T ! horizontal total diffusion Dout(iTxdif) == 12*T ! horizontal XI-diffusion Dout(iTydif) == 12*T ! horizontal ETA-diffusion Dout(iTsdif) == 12*T ! horizontal S-diffusion Dout(iTvdif) == 12*T ! vertical diffusion ! Logical switches (TRUE/FALSE) to activate writing of time-averaged, ! biological processes diagnostics terms into DIAGNOSTIC output file [Ngrids]. Dout(iCOfx) == T ! air-sea CO2 flux Dout(iDNIT) == T ! denitrification flux Dout(ipCO2) == T ! CO2 partial pressure Dout(iO2fx) == T ! air-sea O2 flux Dout(iPPro) == T ! primary production Dout(iNO3u) == T ! NO3 uptakeCode: Select all
DO ivar=1,NDbio2d ifield=iDbio2(ivar) IF (Dout(ifield,ng)) THEN IF (ivar.eq.ipCO2) THEN scale=1.0_r8 ELSE scale=1.0_r8/(REAL(nDIA(ng),r8)*dtBIO) ! mmole m-2 day-1 END IF ... END IF END DO - A new C-preprocessing option FLOAT_STICKY was added to reflect floats that hit the surface and stick floats that hit the bottom. This is needed to avoid overshoots and spurious unmixing near the boundary when vertical random walk (FLOAT_VWALK) is activated. Many thanks to Mark Hadfield for contributing this option.
- A new vertical stretching function, Vstretching = 4, was added and it is denoted as Shchepetkin (2010). This is a major improvement to his 2005 function, Vstretching = 2. The new function, C(s), has a double stretching transformation:
Notice that the bottom function is the second stretching of an already stretched transform. The resulting function is continuous with respect the control parameters theta_s and theta_b with a meaningful range of:
Code: Select all
C(s) = [1 - COSH(theta_s * s)] / [COSH(theta_s) - 1] with bottom refinement: C(s) = [EXP(theta_b * C(s)) - 1] / [1 - EXP(-theta_b)]Due to its functionality and properties Vtransform = 2 and Vstretching = 4 are now the default values for ROMS. Many thanks to Sasha Shchepetkin for designing this nice and continuous transformation.Code: Select all
0 < theta_s <= 10.0 0 <= theta_b <= 4.0
Notice that with Vtransform = 2 the true sigma-coordinate system is recovered as hc goes to infinity. This is useful when configuring applications with flat bathymetry and uniform level thickness. Practically, you can achieve this by setting the following in ocean.in:Please checkCode: Select all
THETA_S = 0.0d0 THETA_B = 0.0d0 TCLINE = 1.0d+17 (a large number) WikiROMS for updated documentation and plots. For example, in the coarse resolution, North Atlantic application (DAMEE_4) we get:
WikiROMS for updated documentation and plots. For example, in the coarse resolution, North Atlantic application (DAMEE_4) we get:
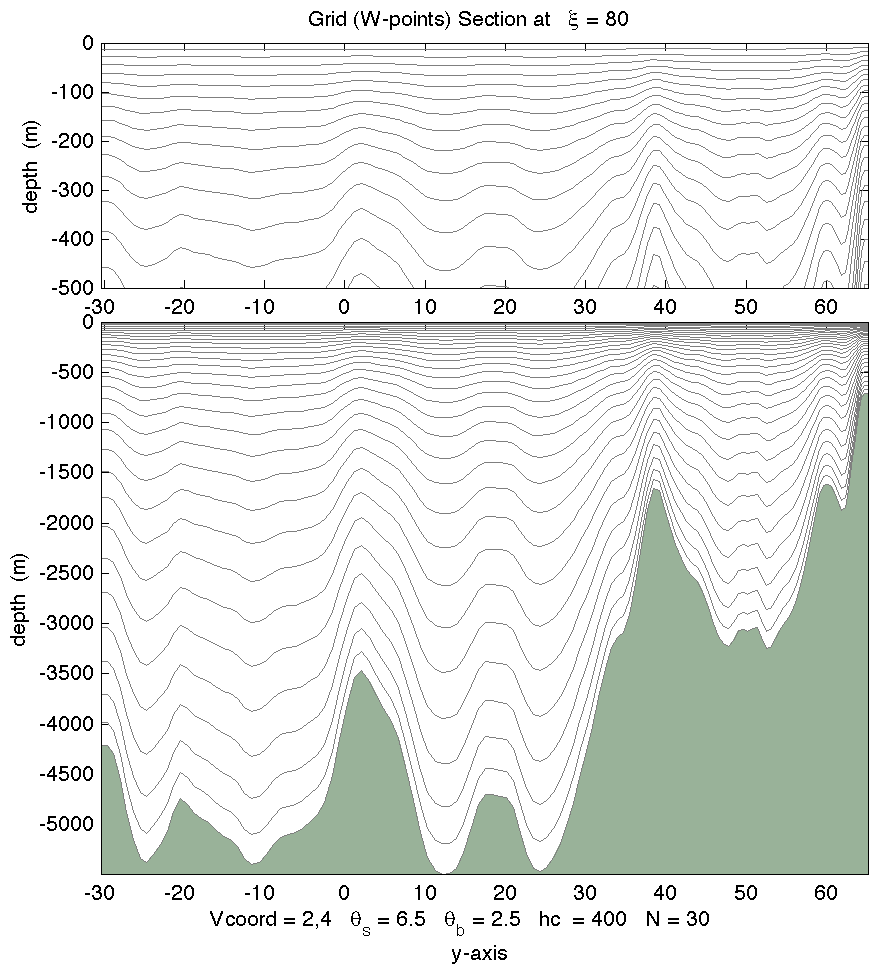
- The following testcases were changed to have the default transformation Vtransform = 2 and Vstretching = 4:
Code: Select all
OLD (1,1) NEW (2,4) theta_s theta_b Tcline theta_s theta_b Tcline --------------------------------------------------------- BASIN 3.3 0.0 1400 3.3 0.0 1400 BENCHMARK 5.0 0.4 200 0.0 0.0 400 BL_TEST 3.2 0.2 5 5.0 1.5 20 CANYON2D 1d-4 0.0 90 0.0 0.0 1d+16 DOUBLE_GYRE 1.0 1.0 50 0.0 0.0 1d+16 FLT_TEST2D 3.0 0.4 100 0.0 0.0 1d+16 FLT_TEST3D 3.0 0.4 100 0.0 0.0 1d+16 GRAV_ADJ 1d-4 0.0 50 0.0 0.0 1d+16 KELVIN 3.0 0.0 50 0.0 0.0 1d+16 LMD_TEST 1d-4 0.0 50 0.0 0.0 1d+16 RIVERPLUME1 3.0 0.4 50 3.0 0.0 30 RIVERPLUME2 3.0 0.4 50 3.0 0.0 30 SEAMOUNT 5.0 0.4 50 6.5 2.0 100 SOLITON 1d-4 0.0 50 0.0 0.0 1d+16 UPWELLING 3.0 0.0 50 3.0 0.0 25 WINDBASIN 1d-4 0.0 50 1.0 0.0 50 - A new driver array_modes_w4dvar.h was added to compute either the requested array mode eigenvector (ARRAY_MODES) or clipped spectrum analysis (CLIPPING) of the stabilized representer matrix during weak constraint, variational data assimilation. The array mode analysis can be used to assess the observing system, prior covariance hypothesis, and the stability of the generalized inverse. The following new CPP options are added:
A new parameter, Nvct, was added to the s4dvar.in input script. If ARRAY_MODES is activated, Nvct eigenvectors of the stabilized representer matrix will be processed (Nvct=1 is the least important, and Nvct=Ninner is the most important). If CLIPPING is activated, only eigenvector Nvct=Ninner will be processed and the rest will be disgarded.
Code: Select all
ARRAY_MODES use if computing W4DVAR representer matrix array modes ARRAY_MODES_SPLIT use to separate analysis due to IC, forcing, and OBC CLIPPING use if computing W4DVAR representer matrix clipping analysis CLIPPING_SPLIT use to separate analysis due to IC, forcing, and OBC RECOMPUTE_4DVAR use if recomputing W4DVAR in analysis algorithms - A new C-preprocessing option, MINRES, was added to accelerate the convergence of the dual formulation data assimilation algorithms (R4D-Var and 4D-PSAS) using the Minimal Residual Method (MinRes) proposed by El Akkraoui and Gauthier (2010, QJRMS, 136, 107-115). This method is described by Paige and Saunders (Sparse Indefinite Systems of Linear Equations, 1975, SIAM Journal on Numerical Analysis, 617-619). Specifically we use equations 6.10 and 6.11 of this paper. This is a remarkable algorithm and makes R4D-Var and 4D-PSAS practical in strong- and weak-constraint applications. The convergence rate is similar to that of the primal form I4D-Var. This is illustrated in the following plot:
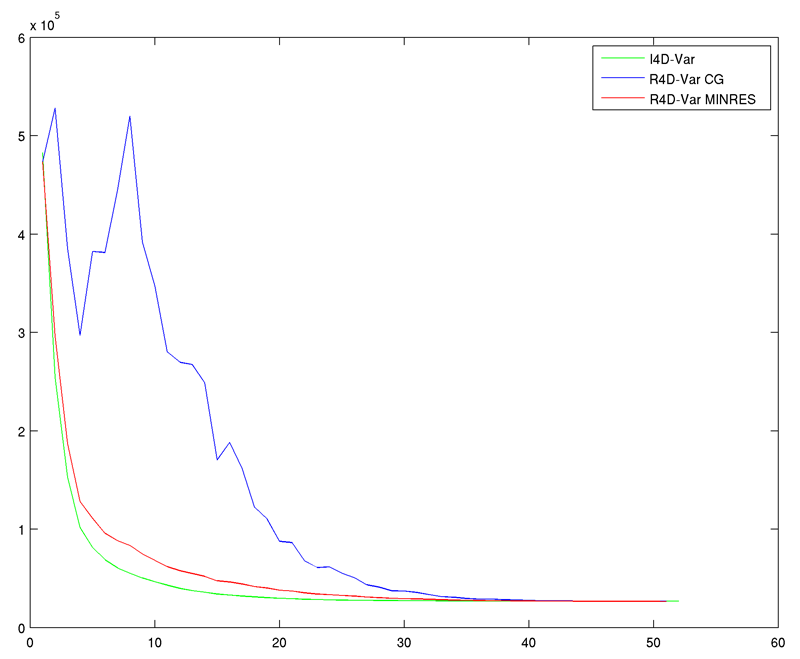
This algorithm also works for the associated observation sensitivity and observation impact. We still need to work on the posterior analysis routines posterior.F and posterior_var.F. These routines will be updated to MINRES later. Many thanks to Andy Moore for his help in coding and testing this new algorithm. Also many thanks to Amal El Akkraoui for bringing this minimization algorithm to our attention at the last International Adjoint Workshop. - All the Generalized Stability Theory (GST), adjoint-based algorithms were updated:
The check-pointing CPP option, CHECKPOINTING, is no longer an internal flag. It is removed from globaldefs.h and the User needs to activate it explicitly in order to create/use the GST restart NetCDF file. Recall the the restart switch LrstGST is activated in the standard input ocean.in file. I removed all the calls to the BLAS library routines daxpy and dnrm2, which is used to computed the Eucledean norm in the GST algorithms. The BLAS library only works in parallel with the PGI compiler. We were getting the wrong norms with the ifort and gfortran compilers. This is due to the legacy code style in this library that dimensions arrays as x(1) instead of x(n). The MPI reduction operations were giving incorrect values. In addition, these routines and others have deprecated arithmetic GO TO and were getting a lot of warnings during compilation. Some of these legacy libraries need to be updated for future compilers that do not support old f77 dimension style. To avoid problems in the future, I wrote my own Eucledean norm routines for real vectors (r_norm2) and complex vectors (c_norm2).
Code: Select all
afte_ocean.h Adjoint Finite Time Eigenmodes (AFT_EIGENMODES) fte_ocean.h Finite Time Eigenmodes (FT_EIGENMODES) fsv_ocean.h Forcing Singular Vectors (FORCING_SV) op_ocean.h Optimal Perturbations (OPT_PERTURBATION) so_semi_ocean.h Stochastic Optimals, Seminorm (SO_SEMI) - The output Ritz vectors from the GST algorithms can be written in separate NetCDF files when the new standard input logical switch LmultiGST is activated:
The eigenvectors for the AFT_EIGENMODES or FT_EIGENMODES are usually complex. In this case, both the real and imaginary eigenvectors are stored in the same file when LmultiGST is activated. Notice that all the eigenvectors have an output record for the initial or final perturbation, except the eigenvectors that have only the adjoint model as a propagator (AFT_EIGENMODES and SO_SEMI). This is a function of the running time window.
Code: Select all
! GST output and check pointing restart parameters. LmultiGST = F ! one eigenvector per file LrstGST = F ! GST restart switch MaxIterGST = 500 ! maximun number of iterations NGST = 10 ! check-pointing interval - The subroutine mp_ncwrite in distributed.F, which used to write 1D/2D state arrays in the GST algorithms, is split into mp_ncwrite1d and mp_ncwrite2d.
- The Forcing Singular Vectors and Stochastic Optimals Vectors are expanded to include the full state (zeta, u, v, temp, salt, ...) and/or surface forcing (sustr, svstr, shflx, ssflux, ...). Several new logical switches were added to the standard input ocean.in to set the desired state vector:
Code: Select all
! Logical switches (TRUE/FALSE) to specify the state variables for ! which Forcing Singular Vectors or Stochastic Optimals is required. Fstate(isFsur) == F ! free-surface Fstate(isUbar) == F ! 2D U-momentum Fstate(isVbar) == F ! 2D V-momentum Fstate(isUvel) == F ! 3D U-momentum Fstate(isVvel) == F ! 3D V-momentum Fstate(isTvar) == F F ! NT tracers Fstate(isUstr) == T ! surface U-stress Fstate(isVstr) == T ! surface V-stress Fstate(isTsur) == F F ! NT surface tracers flux - Added spatially varying bottom roughness length (ZoBot(i,j); m), linear bottom drag coefficients (rdrag(i,j); m/s), or quadratic bottom drag coefficients (rdrag2(i,j); non-dimensional). This can be activated with new C-preprocessing option UV_DRAG_GRID. These spatially varying bottom friction parameters can be specified with either analytical expressions using new C-preprocessing option ANA_DRAG in new routine ana_drag.h or read from application GRID NetCDF file. These parameters are defined at RHO-points and processed according to the momentum bottom friction options UV_LOGDRAG (ZoBot), UV_LDRAG (rdrag), or UV_QDRAG (rdrag2). If reading these variables from the GRID NetCDF, you need to have the following metadata:
Notice that only one of these variables is needed according to the C-preprocessing option UV_LOGDRAG, UV_LDRAG, or UV_QDRAG, respectively. It is also possible to use spatially varying ZoBot when SEDIMENT and/or internal BBL_MODEL options are/is activated. The user is allowed to have different NetCDF variable names for these bottom friction parameters. This can be done in the metadata file varinfo.dat. Any of these time-independent variables are written to the header of output NetCDF files when UV_DRAG_GRID is activated.
Code: Select all
double ZoBot(eta_rho, xi_rho) ; ZoBot:long_name = "time invariant, bottom roughness length" ; ZoBot:units = "meter ; ZoBot:coordinates = "lon_rho lat_rho" ; double rdrag(eta_rho, xi_rho) ; rdrag:long_name = "linear bottom drag coefficient" ; rdrag:units = "meter second-1 ; rdrag:coordinates = "lon_rho lat_rho" ; double rdrag2(eta_rho, xi_rho) ; rdrag2:long_name = "quadratic bottom drag coefficient" ; rdrag2:coordinates = "lon_rho lat_rho" ;
Notice that the sediment variable bottom(i,j,izdef) is initialized to ZoBot. However, bottom(i,j,izdef) changes in time due to sediment processes. There is a difference in these variables due to the time dependency which affect the adjoint model. - The standard output information has been expanded to include the maximum Courant number values and its location. The Courant number in 3D applications is defined at RHO-points as:
The (i,j,k) location where the maximum of C occurs is also reported. Finally, the maximum value of the horizontal speed, SQRT(u*u + v*v), is also reported. This information may be useful to monitor the model CFL condition. It also indicates the places in the grid where the CFL is violated during model blow-up. For example, we get in standard output:
Code: Select all
C = Cu + Cv + Cw where Cu = ABS(u(i,j,k) + u(i+1,j,k)) * dt * pm(i,j) Cv = ABS(v(i,j,k) + v(i,j+1,k)) * dt * pn(i,j) Cw = ABS(wvel(i,j,k-1) + wvel(i,j,k)) * dt * Hz(i,j,k)Notice that this information is written after the global energy and volume diagnostics. The total value of C is not written in 3D applications, but it can be obtained by summing the values of Cu, Cv, and Cw. This information also gives you the clue of which component of C is getting is the largest. In some applications with rotated tensors the value of Cw may get large and the time-step needs to be reduced to avoid the model to blow-up.Code: Select all
STEP Day HH:MM:SS KINETIC_ENRG POTEN_ENRG TOTAL_ENRG NET_VOLUME C => (i,j,k) Cu Cv Cw Max Speed 960 40 00:00:00 8.657897E-04 1.948261E+04 1.948261E+04 2.182663E+17 (096,125,01) 9.919103E-03 4.299619E-03 1.211318E-02 1.049095E+00
The total value of C is reported in 2D application, since we have an information column available (no Cw value is possible):Code: Select all
STEP Day HH:MM:SS KINETIC_ENRG POTEN_ENRG TOTAL_ENRG NET_VOLUME C => (i,j,k) Cu Cv C Max Max Speed - Added the capability for writing surface pressure (Pair) and surface wind components (Uair, Vair) to history, averages, and station output NetCDF files. This is done for convenience during post-processing of the model output. For example, it can be used to plot the dependency between surface elevation, pressure, and winds at a particular station location. The winds are written as provided at input, ROMS curvilinear coordinates (XI, ETA) and rotated angles, if applicable. This can be activated in the standard input file ocean.in by turning on the following new switches:
and station.in input script:
Code: Select all
Hout(idPair) == T ! surface air pressure Hout(idUair) == T ! surface U-wind component Hout(idVair) == T ! surface V-wind component ... Aout(idPair) == T ! surface air pressure Aout(idUair) == T ! surface U-wind component Aout(idVair) == T ! surface V-wind componentCode: Select all
Sout(idPair) == T ! surface air pressure Sout(idUair) == T ! surface U-wind component Sout(idVair) == T ! surface V-wind component